# Proteomics analysis of Gargling samples from CoviD-19 infected patients
# Live Resources
| usegalaxy.eu |
|---|
# Description
Ihling et al present a protein MS-based ‘proof-of-principle’ method to detect SARS-CoV-2 virus proteins from gargle samples from COVID-19 patients. Their protocol consists of an acetone precipitation step, followed by tryptic digestion of gargle solution proteins, followed by MS analysis. In the original manuscript, the authors detect peptides from SARS-CoV-2 virus proteins and present evidence for their spectral annotation. This study is an initiative in developing a routine MS-based diagnostic method for COVID-19 patients.
# Workflow
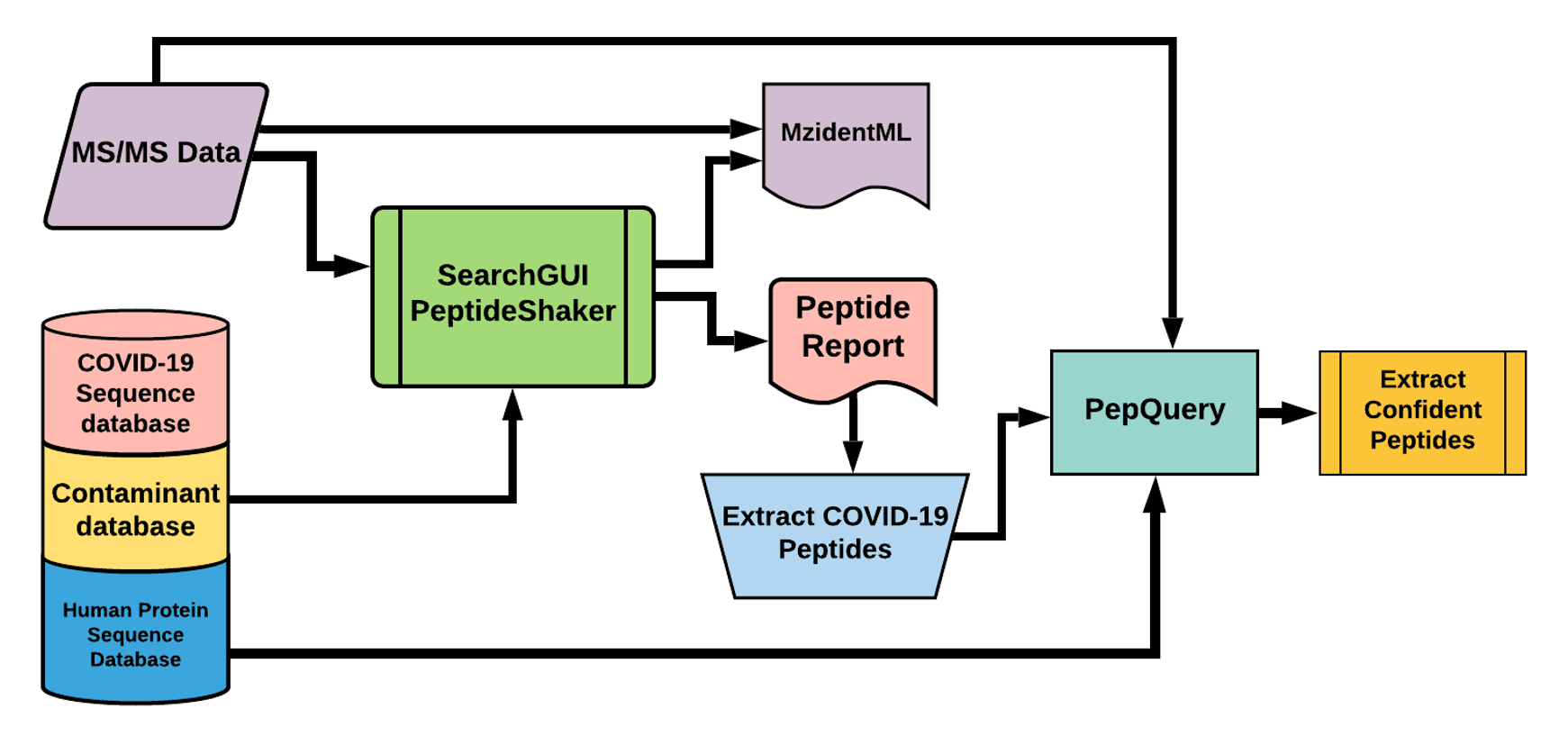
The Galaxy workflow includes RAW data conversion to MGF and mzML format. The MGF files are searched against the combined database of Human Uniprot proteome, contaminant proteins and SARS-Cov-2 proteins database using X!tandem, MSGF+, OMSSA search algorithms with SearchGUI and FDR and protein grouping using PeptideShaker. This resulted in detection of nine peptides from SARS-CoV-2 proteins.
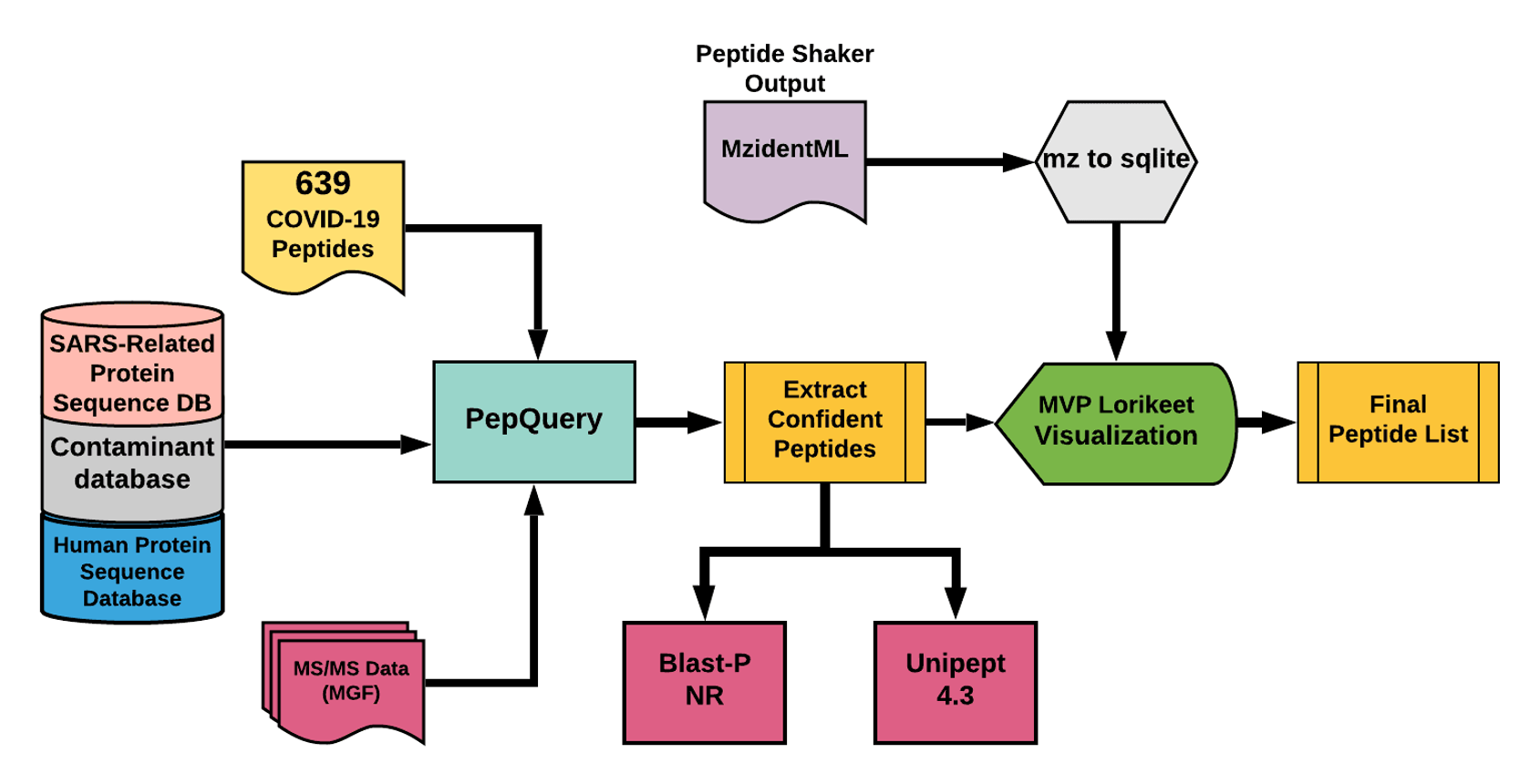 The detected peptides were searched against NCBInr to ascertain that these peptides were specific to SARS-CoV-2 proteins. Also, the detected peptides were subjected to analysis by PepQuery and Lorikeet to ascertain the quality of peptide identification.
The detected peptides were searched against NCBInr to ascertain that these peptides were specific to SARS-CoV-2 proteins. Also, the detected peptides were subjected to analysis by PepQuery and Lorikeet to ascertain the quality of peptide identification.
# Results
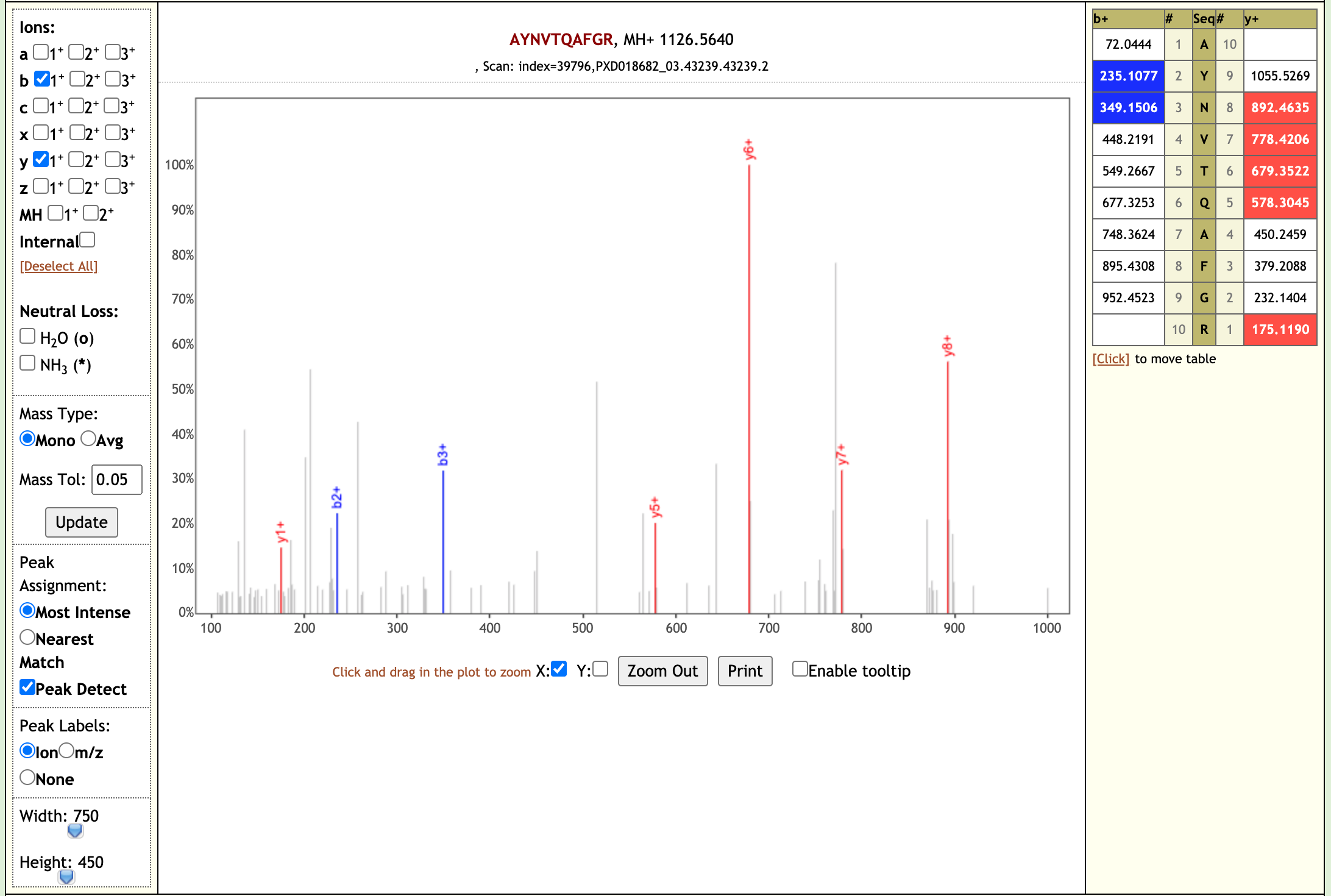
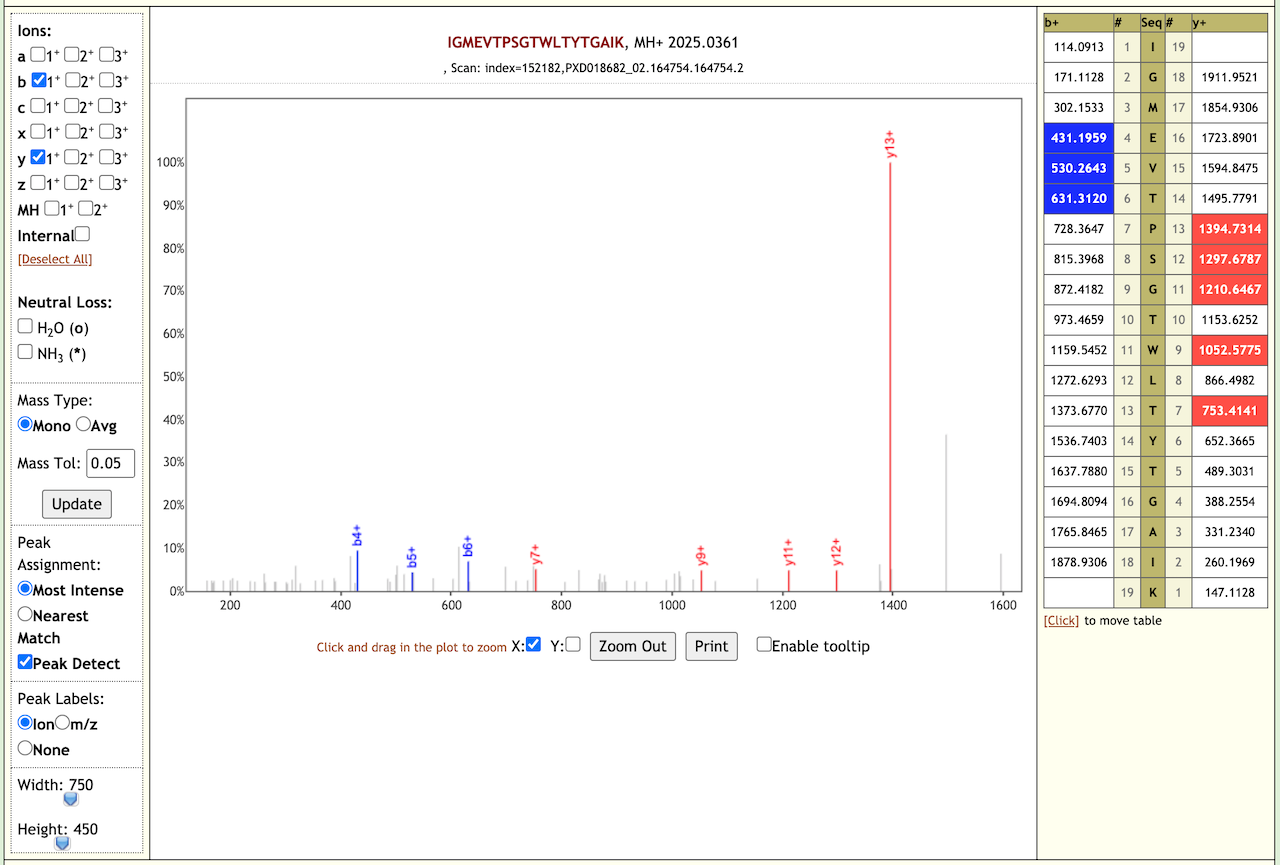
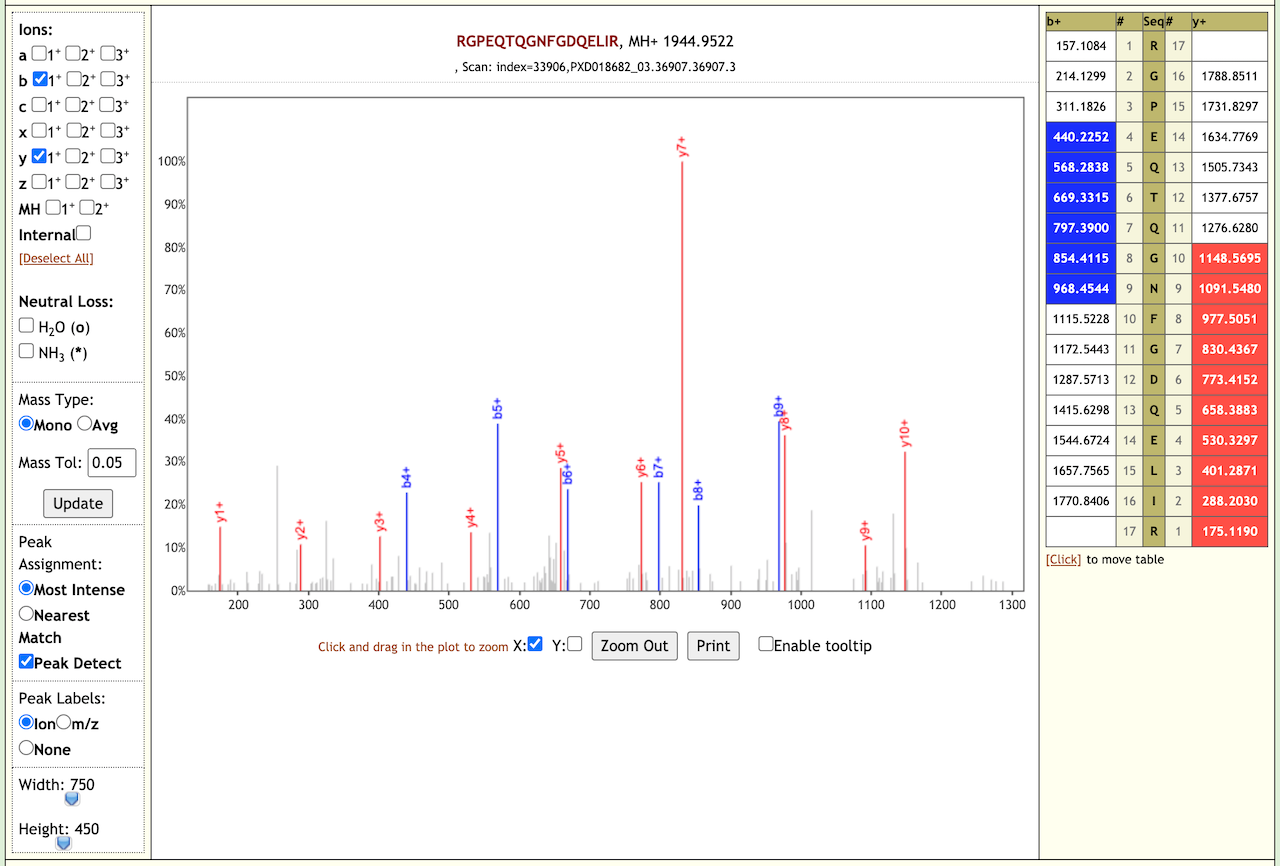
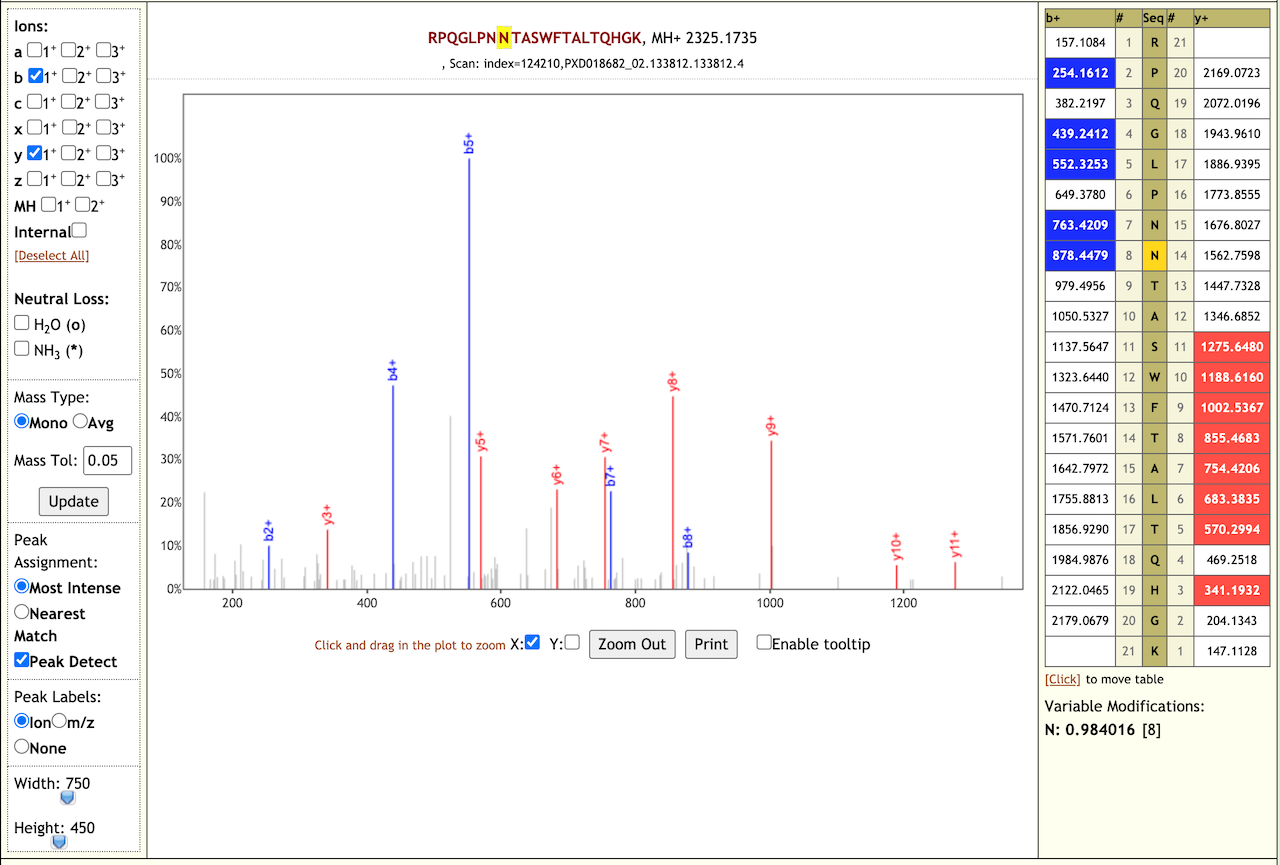
The database search workflow yielded us 8 COV-2 peptides from the second and third raw files.
When we performed the validation search using PepQuery it gave us 21 COV-2 peptides. We also checked the Lorikeet spectra of these peptides. Here are some examples of the Lorikeet spectra using the Multiomics visualization tool in the Galaxy platform.