# Proteomic re-analysis of nasopharyngeal swab samples from COVID-19 patients
# Live Resources
| usegalaxy.eu |
|---|
# Description
Bankar et.al (opens new window) performed proteomics study on nasopharyngeal swab samples from COVID-19 patients sample. This study was performed to identify a panel of host proteins which could serve as biomarkers of severity prediction .Data-dependent acquisition MS spectra were acquired using Orbitrap-Fusion mass spectrometer.
# Workflow
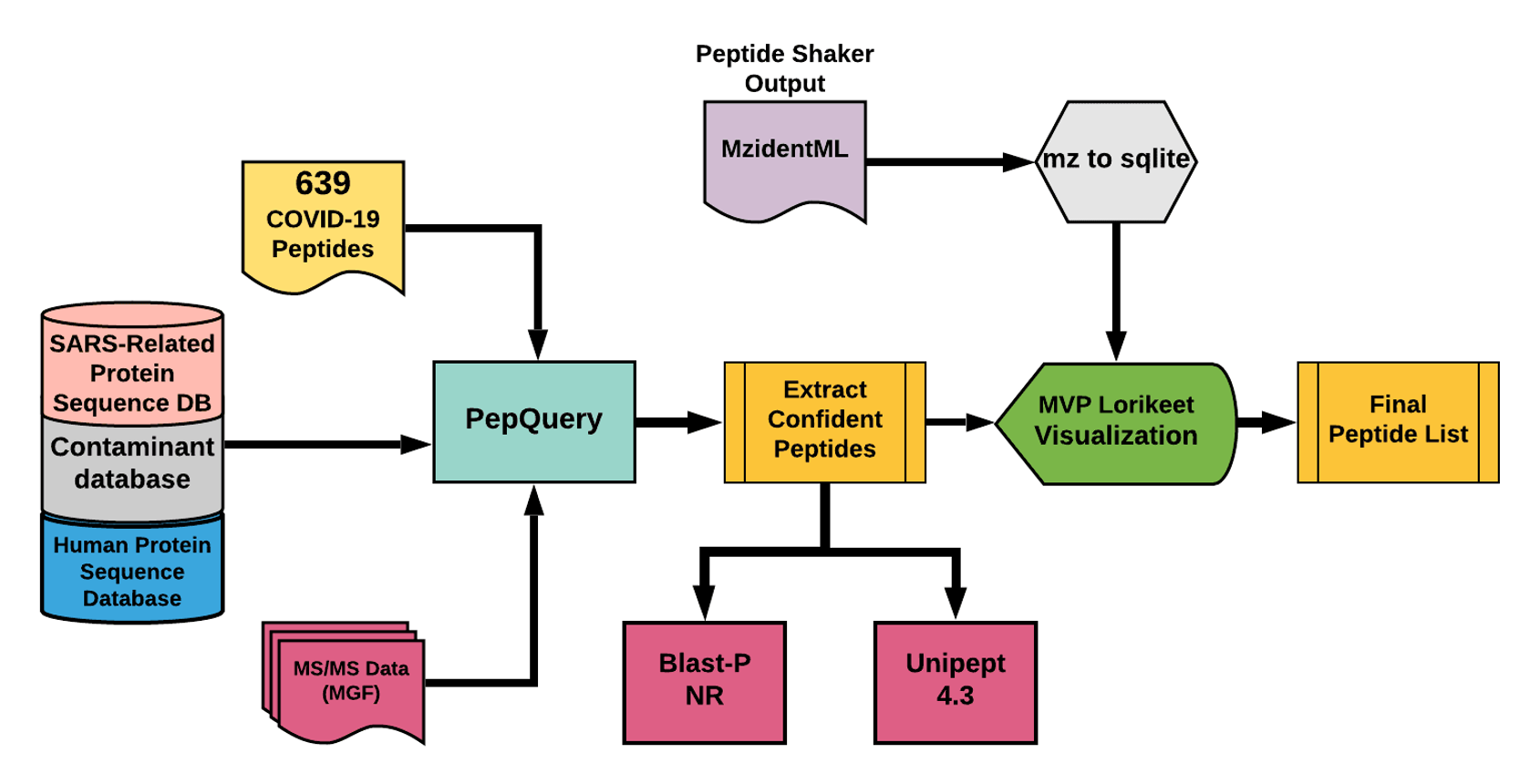
The Galaxy workflow includes RAW data conversion to MGF and mzML format. The MGF files are searched against the combined database of Human Uniprot proteome, contaminant proteins and SARS-Cov-2 proteins database using PepQuery Validation workflow. This resulted in detection of 35 peptides from SARS-CoV-2 proteins. The detected peptides were searched against NCBInr to ascertain that these peptides were specific to SARS-CoV-2 proteins. The detected peptides were later subjected to analysis by Lorikeet visualization to ascertain the quality of peptide identification.
# Results
We detected 35 COVID-19 peptides in the nasopharyngeal swab samples. The peptides were subjected to BLAST-P and Lorikeet analysis to ascertain the validity of peptide spectral matches. Our results validated the presence of peptide "DGIIWVATEGALNTPK" with good spectral quality.