# Proteomics analysis of naso-pharyngeal swabs samples from COVID-19 infected and non-infected individuals
# Live Resources
| usegalaxy.eu |
|---|
# Description
Rivera et al (opens new window) performed comparative quantitative proteomic analysis from oro- and naso-pharyngeal swabs used for COVID-19 diagnosis. Tryptic peptides obtained from five COVID-19 positive and five COVID-19 negative samples were analysed by LC-MS/MS using a Q-Exactive Plus mass spectrometer. The mass spectrometry (MS) data was made available via ProteomeXchange (PXD020394 (opens new window)) so as to facilitate the use of MS-based approaches for COVID-19 diagnosis.
# Workflow
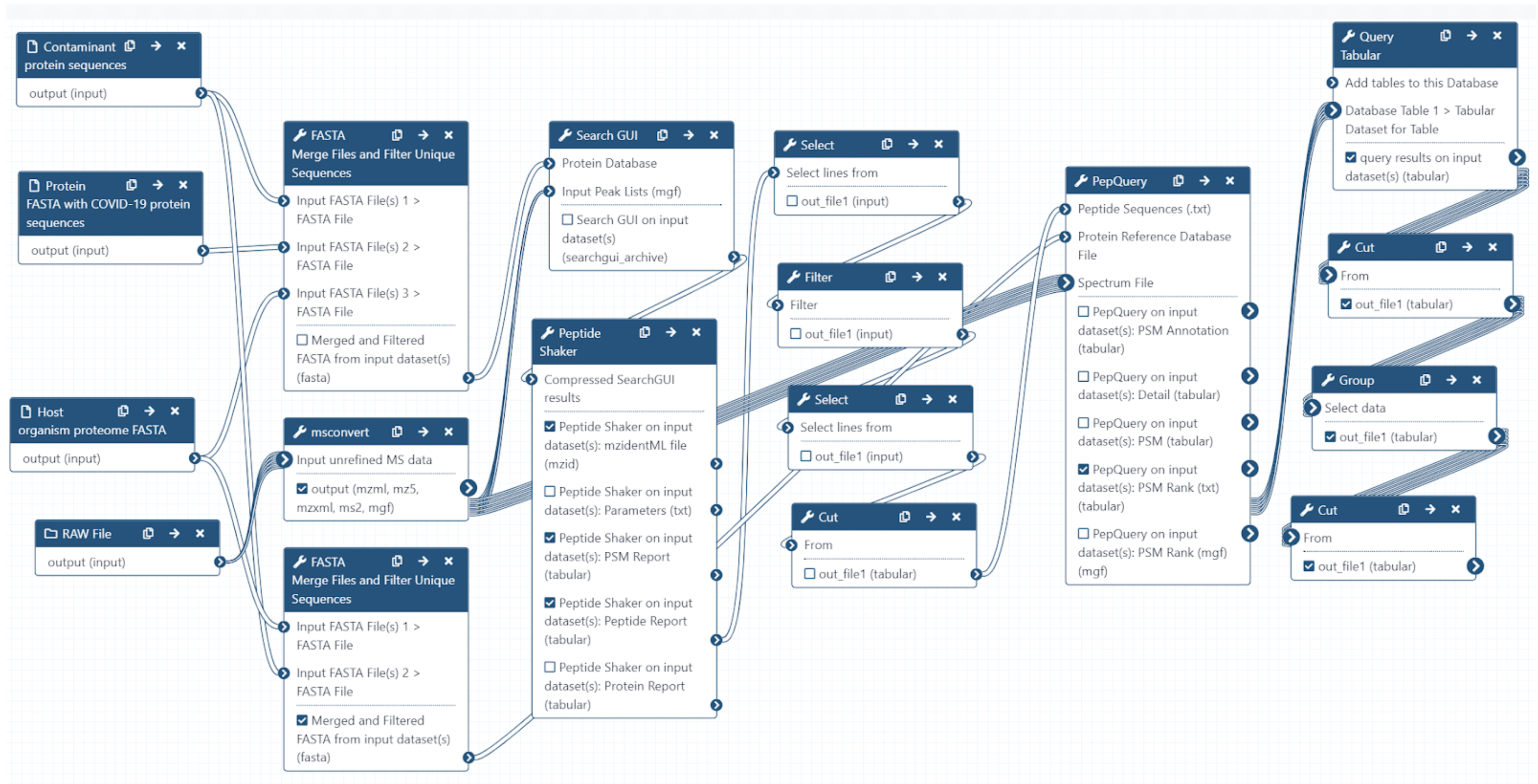
The Galaxy workflow includes RAW data conversion to MGF and mzML format. The MGF files are searched against the combined database of Human Uniprot proteome, contaminant proteins and SARS-Cov-2 proteins database using X!tandem, MSGF+, OMSSA search algorithms with SearchGUI and FDR and protein grouping using PeptideShaker. This resulted in detection of ten peptides from SARS-CoV-2 proteins.
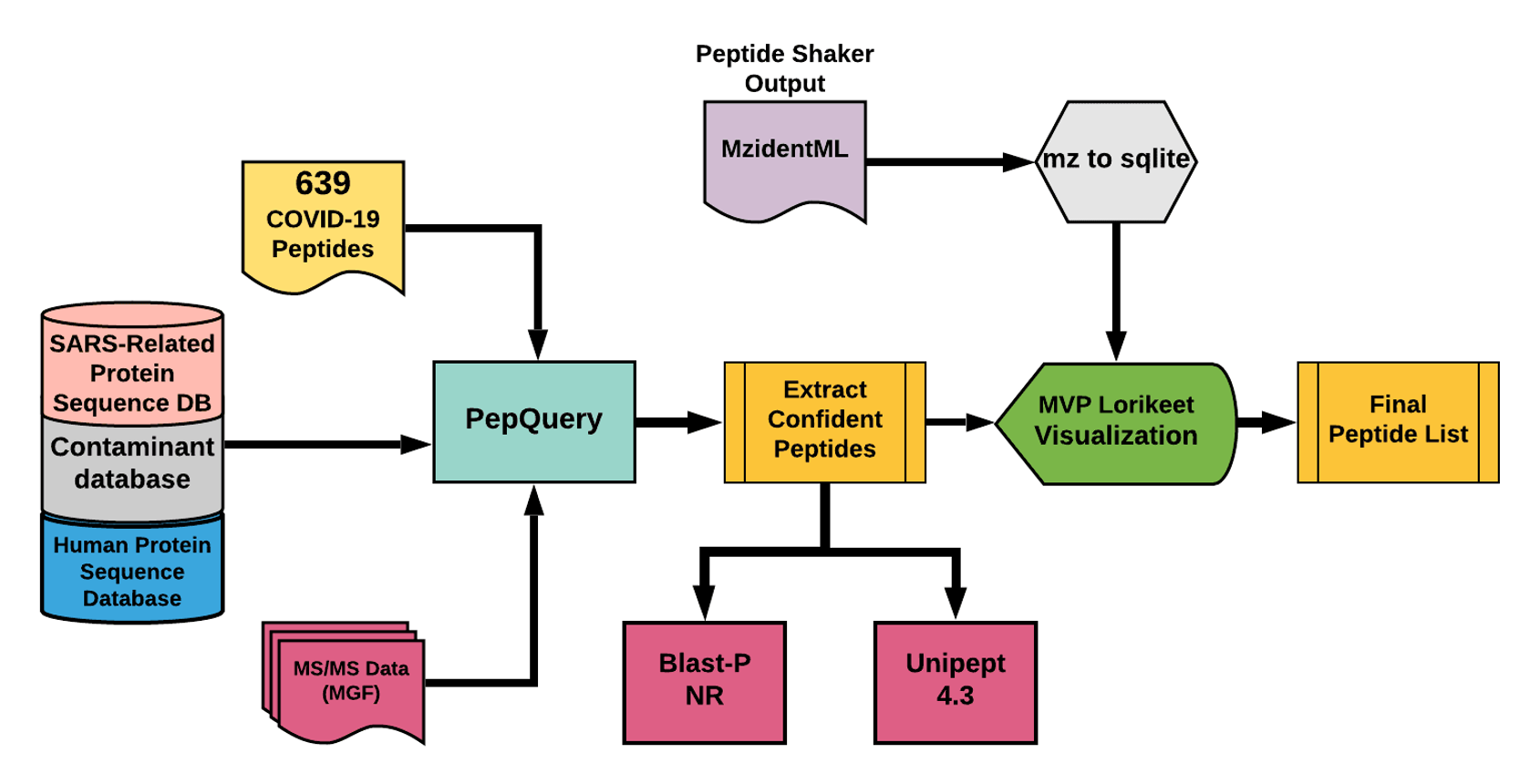
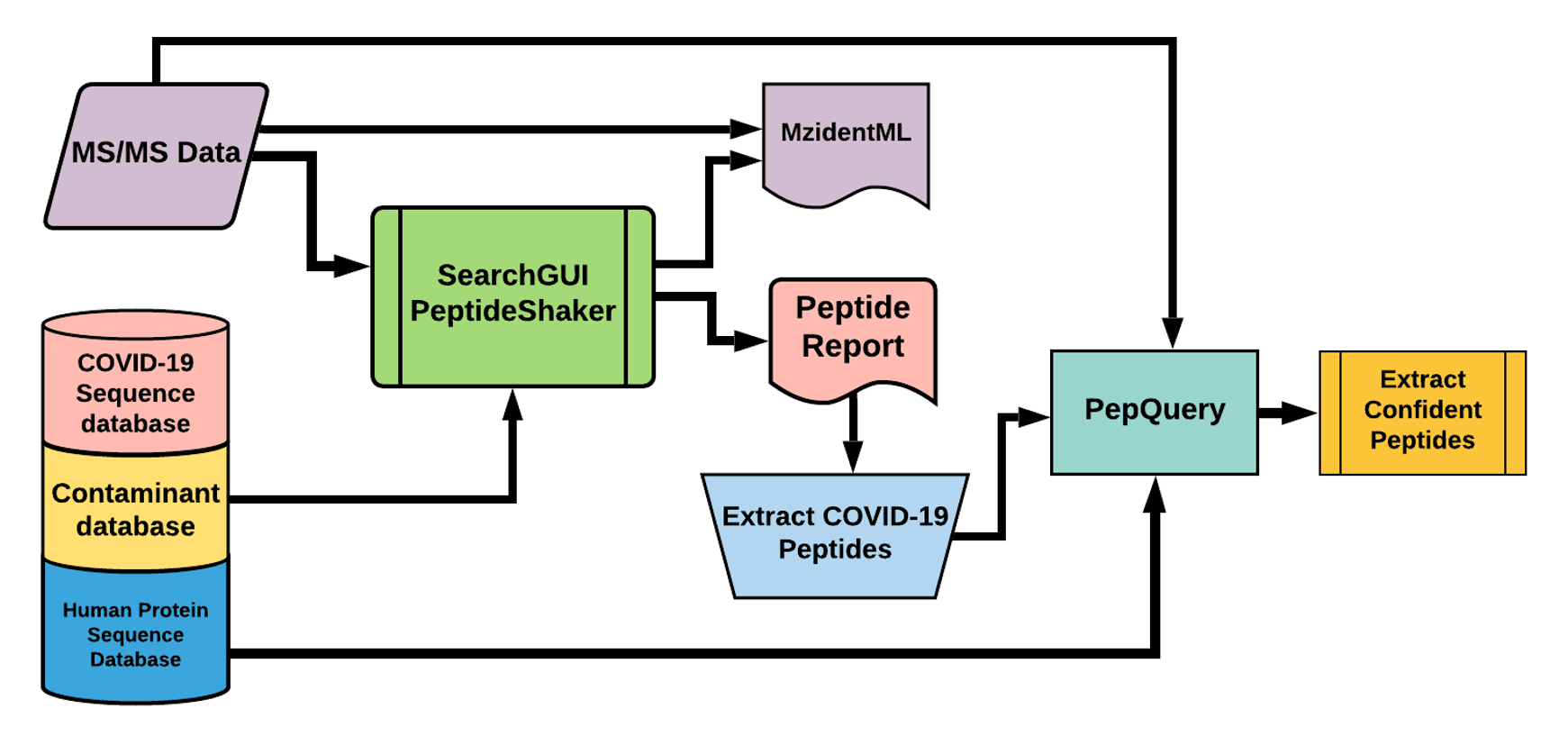 The detected peptides were searched against NCBInr to ascertain that these peptides were specific to SARS-CoV-2 proteins.
The detected peptides were later subjected to analysis by PepQuery and Lorikeet to ascertain the quality of peptide identification.
The detected peptides were searched against NCBInr to ascertain that these peptides were specific to SARS-CoV-2 proteins.
The detected peptides were later subjected to analysis by PepQuery and Lorikeet to ascertain the quality of peptide identification.
# Results
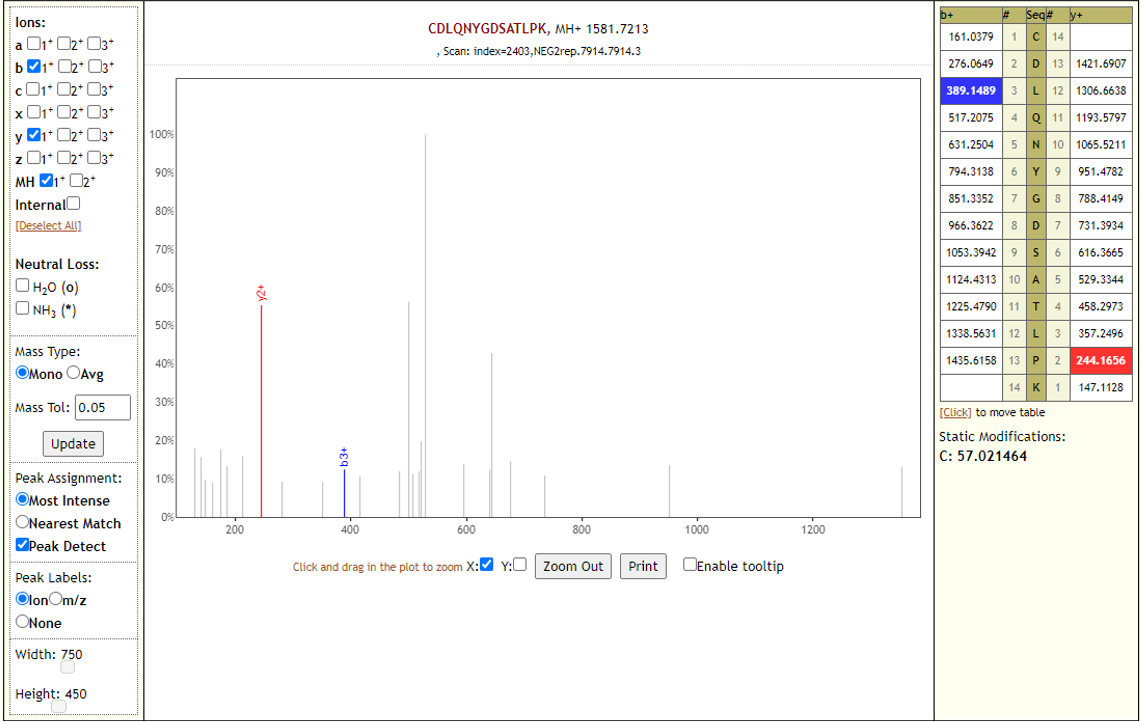
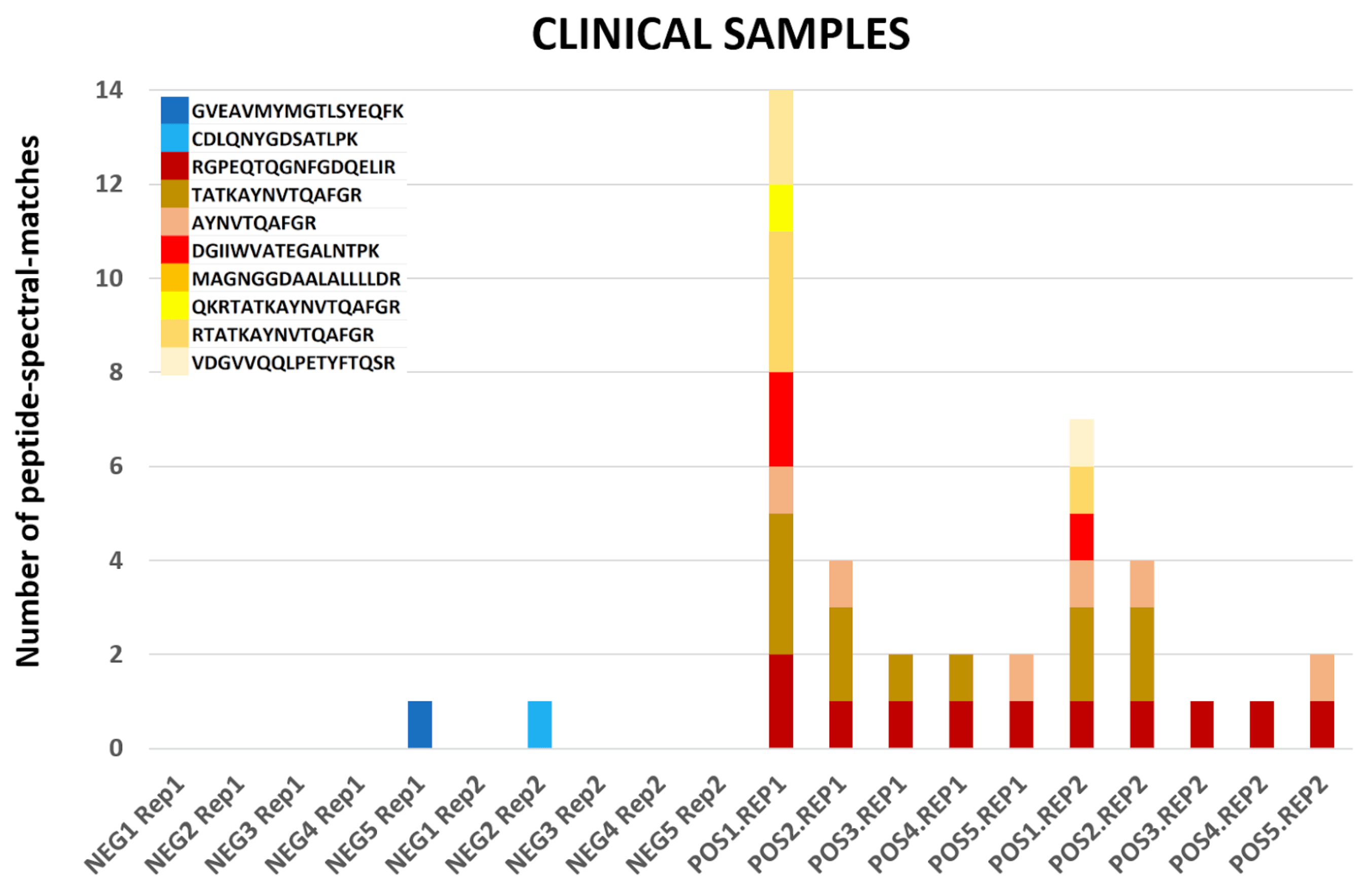 The COVID-19 positive patient samples detected peptides from SARS-CoV-2 proteins (see below). The Peptide search workflow yields 9 COV-2 peptides and the Peptide Validation workflow yields 10 COV-2 peptides.
The COVID-19 positive patient samples detected peptides from SARS-CoV-2 proteins (see below). The Peptide search workflow yields 9 COV-2 peptides and the Peptide Validation workflow yields 10 COV-2 peptides.
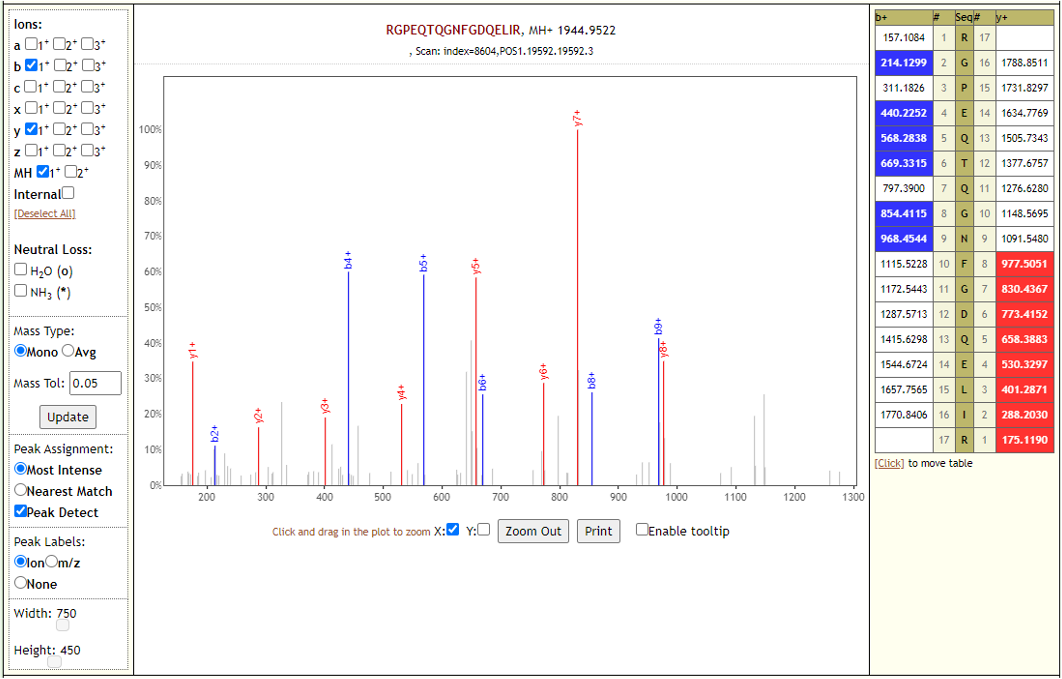
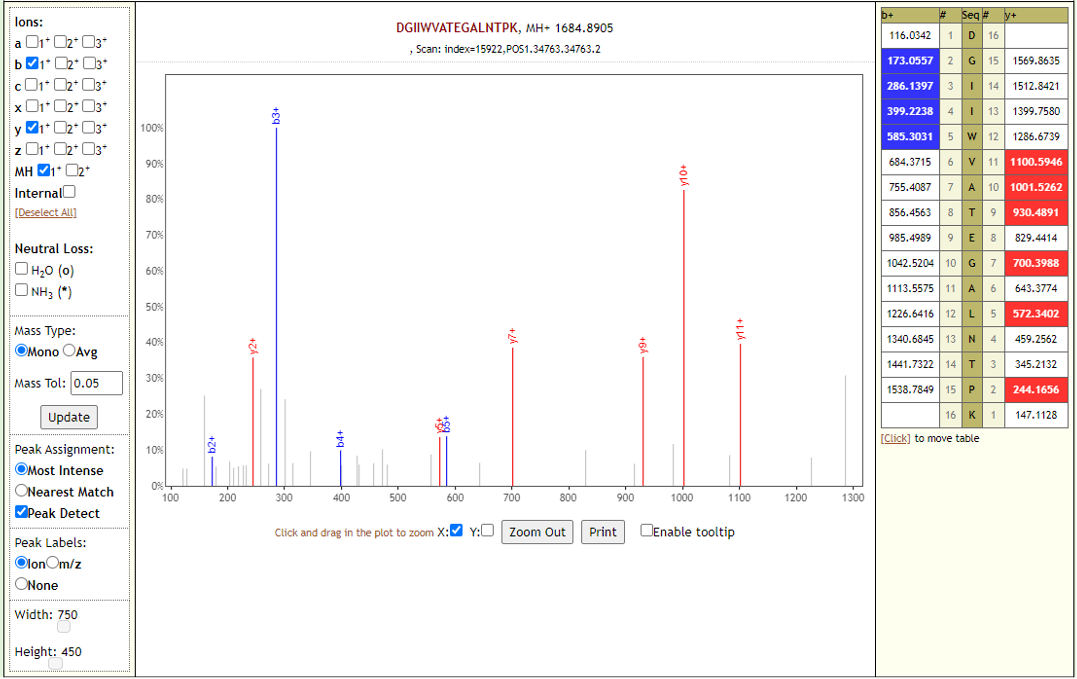
COVID-19 negative samples also detected two peptides from COVID-19 proteins - however Lorikeet analysis showed that the spectral evidence for these peptides did not support the peptide-spectral-match as shown below.