# Assembly of SARS-CoV-2 from pre-processed reads
# Live Resources
| usegalaxy.org | usegalaxy.eu | usegalaxy.org.au | usegalaxy.be | usegalaxy.fr |
|---|---|---|---|---|
# What's the point?
Use a combination of Illumina and Oxford Nanopore reads to produce SARS-CoV-2 genome assembly.
# Outline
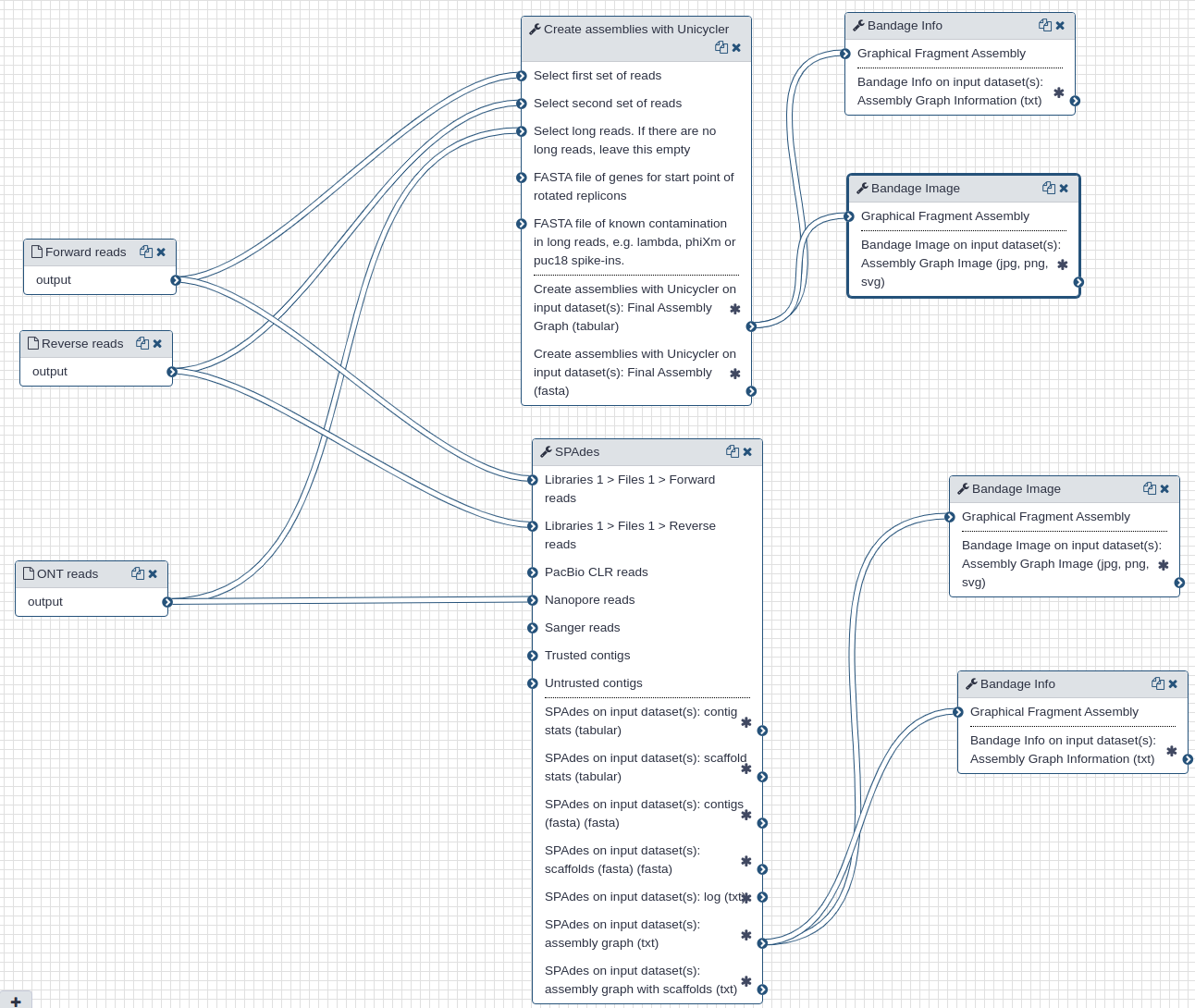
We use Illumina and Oxford Nanopore reads that were pre-processed to remove human-derived sequences. We use two assembly tools: spades (opens new window) and unicycler (opens new window). While spades is a tool fully dedicated to assembly, unicycler is a "wrapper" that combines multiple existing tools. It uses spades as an engine for short read assembly while utilizing mimiasm (opens new window) and racon (opens new window) for assembly of long noisy reads.
In addition to assemblies (actual sequences) the two tools produce assembly graphs that can be used for visualization of assembly with bandage (opens new window).
# Inputs
Filtered Illumina and Oxford Nanopore reads produced during the pre-processing step are used as inputs to the assembly tools.
# Outputs
Each tool produces assembly (contigs) and assembly graph representations. The largest contigs generated by unicycler and spades were 29,781 and 29,907 nts, respectively, and had 100% identity over their entire length.
The following figures show visualizations of assembly graphs produced with spades and unicycler. The complexity of the graphs is not surprising given the metagenomic nature of the underlying samples.
| Assembly graphs for Unicycler (A) and SPAdes (B) |
|---|
| A. Unicycler assembly graph |
| B. SPAdes assembly graph |
# History and workflow
A Galaxy workspace (history) containing the most current analysis can be imported from here (opens new window).
The publicly accessible workflow (opens new window) can be downloaded and installed on any Galaxy instance. It contains version information for all tools used in this analysis.
# BioConda
Tools used in this analysis are also available from BioConda:
| Name | Link |
|---|---|
unicycler | |
spades | |
bandage |