| usegalaxy.eu |
|---|
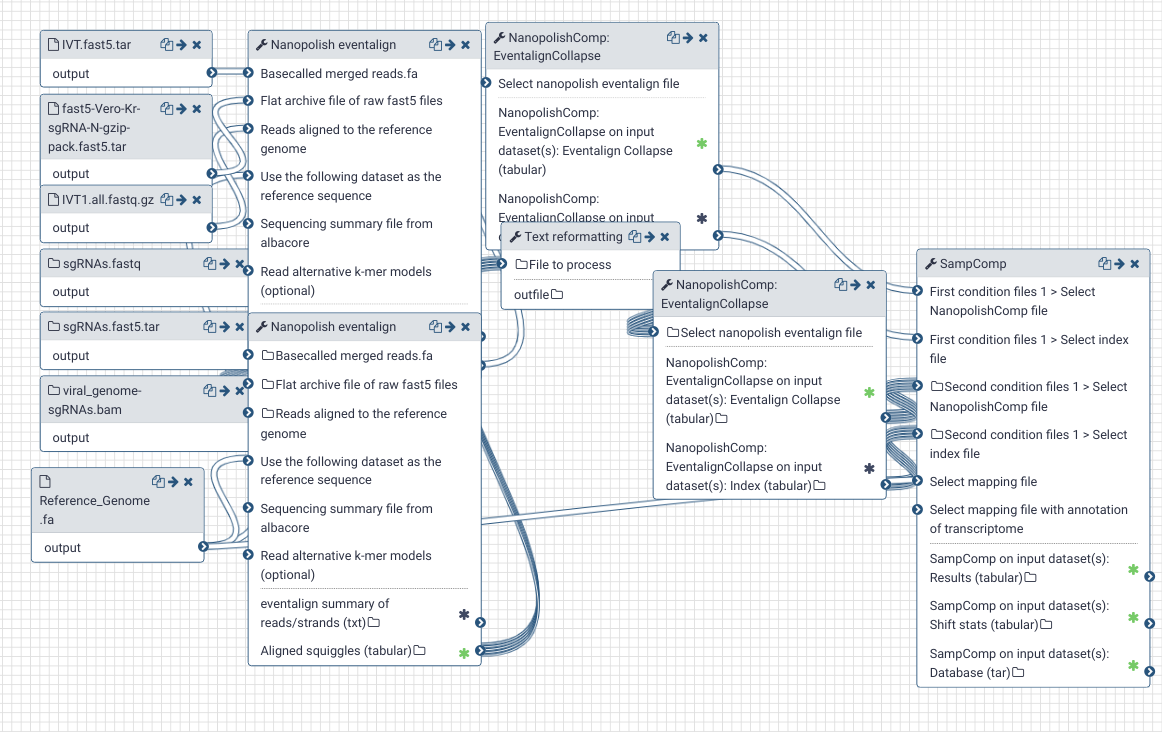
# Methylation analysis
The most reliable approach for identifying the putatively modified nucleotides from the sequencing data is by comparing the distribution of raw electrical signals between two conditions. Tombo and Nanocompore are two tools that can be used for this purpose. Here, as the alternative condition, we use the in vitro transcribed oligos of SARS-CoV-2 from Kim et al..