# Scoring
This section describes how the docking poses were scored. Whilst docking does generate binding scores, these are widely known not to correlate well with binding affinities, especially for fragments, so two additional tools developed at Oxford University were used to generate additional predicted binding scores.
# Live Resources
| usegalaxy.eu |
|---|
# Outline
- The 25 docked poses for each enumerated charge state of the candidate compounds were scored using TransFS (opens new window) [1]. This step
is executed in a Docker container and requires a GPU for execution. Scores range from 0 (bad) to 1 (good). The score is
recorded as the
TransFSScoreproperty in the resulting SD file. - Those scored poses are sorted and filtered so that only the top TransFS scoring pose for each original candidate (before charge enumeration) is retained.
- Those top poses are then scored with the SuCOS Max (opens new window) [2] tool to generate a score that reflects the charge and feature overlap with the original 17 fragment ligands. Bigger scores are better and indicate more overlap.
The resulting poses have scores from rDock for docking as well as for TransFS (opens new window) and SuCOS (opens new window).
# Histories and workflows
A Galaxy workspace (history) containing the most current analysis can be imported from here (opens new window).
The publicly accessible TransFS scoring workflow (opens new window) and SuCOS scoring workflow (opens new window) can be downloaded and installed on any Galaxy instance. They contain version information for all tools used in this analysis.
| Scoring workfows |
|---|
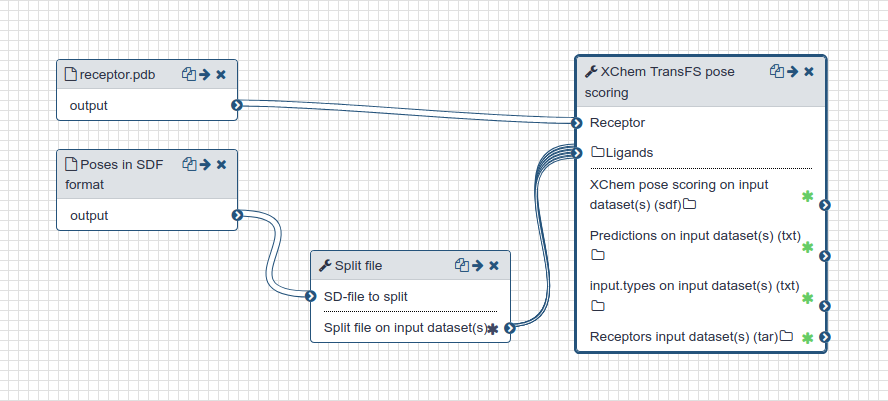 |
| TransFS scoring workflow |
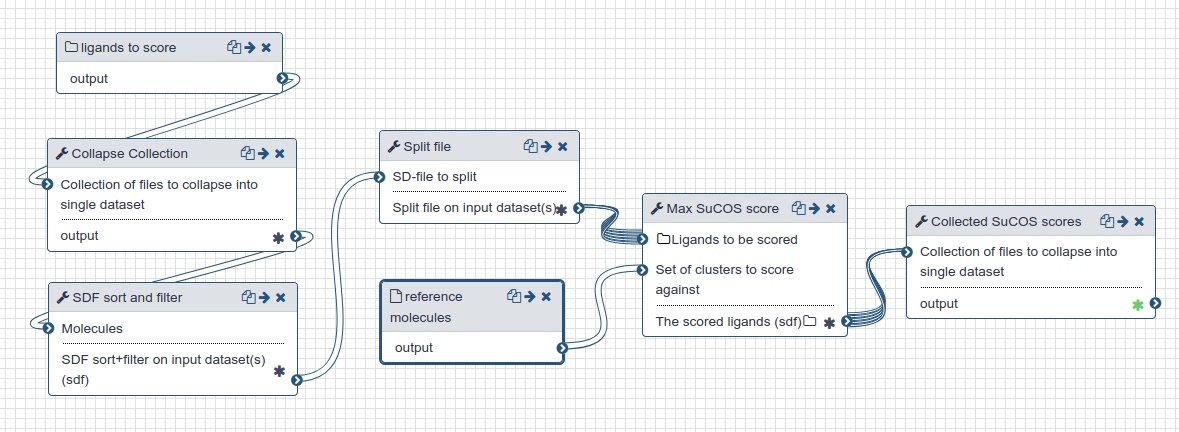 |
| SuCOS scoring workflow |
# References
[1] TransFS -Scantlebury et al., Dataset Augmentation Allows Deep Learning-Based Virtual Screening To Better Generalise To Unseen Target Classes, And Highlight Important Binding Interactions. URL: https://www.biorxiv.org/content/10.1101/2020.03.06.979625v1
[2] SuCOS - Leung et al., SuCOS is Better than RMSD for Evaluating Fragment Elaboration and Docking Poses, doi:10.26434/chemrxiv.8100203.v1 (opens new window)