# Preparation for docking
This section describes the preparation of protein and ligands for docking.
# Live Resources
| usegalaxy.eu |
|---|
# Outline
- The PDB files from crystallography were prepared by removing all crystallographic waters and then protonated at pH 7.4 using OpenBabel.
- The initial ~42,000 candidate compounds in SMILES format are enumerated using Dimorphite-DL [1] and RDKit (opens new window) to a total of ~150,000 compounds with relevant charge forms.
- For each of these, a three-dimensional structure is generated using OpenBabel [2] and saved in SD-format.
- The SD-file is split into chunks of 1,000 molecules each ready for docking.
# History and workflow
A Galaxy workspace (history) containing the most current analysis can be imported from here (opens new window).
The publicly accessible workflow (opens new window) can be downloaded and installed on any Galaxy instance. It contains version information for all tools used in this analysis.
| Preparation for docking |
|---|
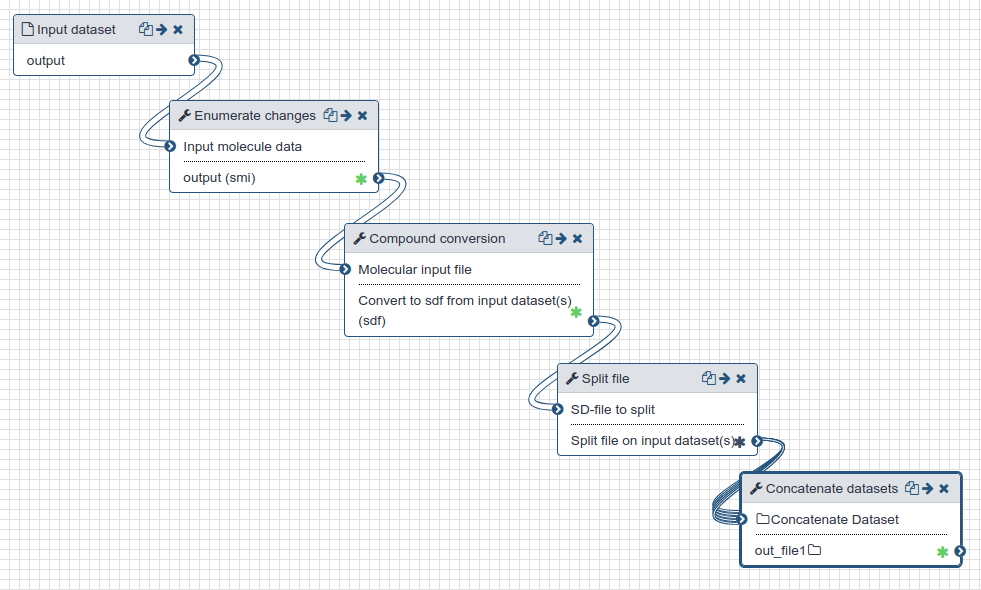 |
| Preparation of protein and ligands for docking. |
# References
[1] Dimorphite-DL: an open-source program for enumerating the ionization states of drug-like small molecules, Robb et al. doi:10.1186/s13321-019-0336-9 (opens new window).
[2] Open Babel: An open chemical toolbox. O'Boyle et. al doi:10.1186/1758-2946-3-33 (opens new window).